A few weeks ago, I had the opportunity to attend a lecture by microbiologist Phillip Aldridge, of the University of Newcastle. The topic of his lecture was “The Regulation of Flagellar Assembly”. Being an ID proponent, I had a natural interest in what Aldridge was going to say, and I had been looking forward to the event for some time. I was already familiar to a degree with several of the key mechanisms and regulation of flagellar biosynthesis. Nonetheless, the lecture succeeded in re-kindling my passion for biology, and inspired me to do some in-depth research on my own with regards the workings of this engineering marvel.
I must confess that I was blown away. If one thought that the functional-specificity of arrangement with respect to the flagellum’s key components may well provide adequate grounds for a design inference, the mechanisms of flagellar construction take this intuition to a whole new level. So mesmerized I was by the motor’s intrinsic beauty and elegance, that I decided to provide a sketch overview of this amazing process for the benefit of readers of this blog. Of course, there are variations in the flagellum’s overall construct from species to species. The archetypical flagellum, however, is probably that of the closely related species, Escherichia coli, Salmonella enterica, and Salmonella typhimurium. It is this that I want to primarily focus on.
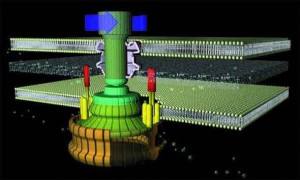
An overview of the flagellum’s construct
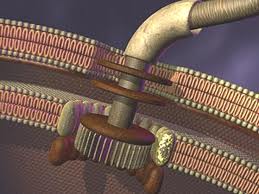
As with a typical man-made motor, the unit which provides rotary motion (which is embedded in the inner cell membrane) revolves within a stationary component (stator). The rotor is a tube-like structure which extends from within the cell, through the membrane, to the outside, where the flagellar filament (which serves as a propeller) is attached via a flexible joint.
The flagellum consists of a basal body (which is embedded in the cell wall) and two axial structures, the book and filament. The basal body is made up of the MS ring, rod, and L- and P- rings. Parts of the axial structure are exported from the cell by the type III secretion system (T3SS). The T3SS is composed of several proteins from the MS ring and a peripheral hexameric ATPase FliI that drives the export process.
The motor is anchored in the cytoplasmic membrane and cell wall. The motor consists of a central rod that passes through a series of rings. In gram-negative bacteria, an outer ring (the L ring) is anchored in the lipopolysaccharide layer. Another ring (the P ring) is anchored in the peptidoglycan layer of the cell wall. A third set of rings (the MS and C rings) are respectively found within the cytoplasmic membrane and the cytoplasm. Gram positive bacteria, by contrast, do not have an outer membrane, and thus only possess the inner pair of rings. A series of proteins, called Mot proteins, surround the inner ring and are anchored in the cytoplasmic membrane. The structures of the L, P, C and MS rings, in combination with the central rod, are collectively referred to as the “basal body”. In addition, a class of proteins called the Fli proteins function as a ‘switch’ for the reversal of motor rotation, which is plugged into an elegant signal transduction mechanism for receiving feedback from the environment — I will discuss this further in due course.
The flagellar filament (propeller) is constructed from subunits of the polypeptide flagellin. There is a wider region at the base of the filament (the ‘hook region’), which connects the filament to the motor component of the base. These subunits are arranged in a tight helical structure to produce a stiff hollow rod. Each subunit contributes to a small propeller blade to the outside of the structure.
The driving force behind the flagellum’s rotary motion comes from a proton motive force, to which I shall return in due course.
Chemotaxis and Signal Transduction
Chemotaxis refers to the means by which bacteria direct their movements in response to environmental chemical signals. This mechanism allows the bacteria to locate food by swimming towards the highest concentration of food molecules (such as glucose). The default direction of flagellar rotation is anticlockwise. This directionality of rotation can be reversed by virtue of a classic two-component signal transduction circuit (discussed below). When the direction of rotation is switched to clockwise, the flagellar bundle breaks apart, and the bacterium physically “tumbles” in place. The overall movement of a bacteria can be described in terms of a series of alternating swimming and ‘tumbling’ phases. When the bacterial cell ‘tumbles’, it re-orientates itself. By repeatedly evaluating its course, the bacterium is able to find favourable locations with the highest concentration of attractant (possibly a food source).
Before we can properly appreciate the details and technicalities of this system, it is necessary to take a step back and understand the foundational principles upon which it is based. Bacteria are able to move towards a food source, such as glucose, by a process known as “chemotaxis.” A requisite for this process to work is the ability of the bacterial flagellar motor to literally shift gears so that it switches from spinning counter-clockwise to rotating clockwise. This change in rotation is brought about in response to chemical stimuli from the cell’s exterior. These chemical signals are detected by a two-component signal transduction circuit that operates to induce the switch in flagellar rotation.
In general, a two-component regulatory system comprises an integral membrane protein known as a “histidine protein kinase,” and a cytoplasmic protein known as a “response regulator.”
The histidine protein kinase has two domains: an input domain and a transmitter domain. The former is located on the outside of the cell, and is ideally situated to detect incoming environmental signals. The latter is situated on the cytoplasmic face of the cell membrane, and is positioned such that it can interact with the response regulator.
An external stimulus causes a conformational change in the histidine protein kinase. This causes the transfer of phosphoryl groups (autophosphorylation) from ATP to a conserved histidine residue. This phosopho-group is then moved to an aspartate residue of the response regulator. This enables the response regulator to bind to the DNA in order to regulate the transcription of its target genes.
What I have thus far described represents a very basic two-component regulatory system. It was, however, necessary to look at the system in principle before we describe its application in the case of chemotaxis.
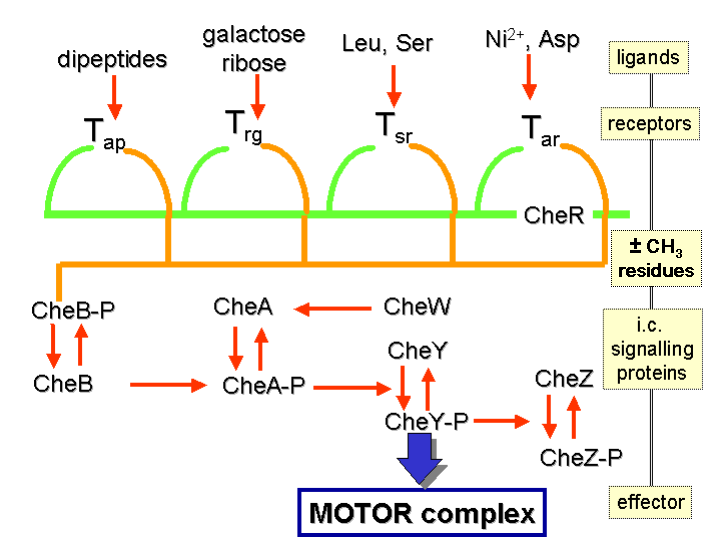
How do bacteria detect a chemical gradient? The answer lies in a certain class of transmembrane receptors called methyl-accepting chemotaxis proteins (hereafter, MCPs). Different MCPs can detect different types of molecules, and are able to bind attractants or repellents. These receptors then communicate with — and activate — the so-called “Che proteins”.
Proteins called CheA and CheW are bound to the receptor. The former is the histidine kinase for this system. Upon activation of the receptor, the CheA’s conserved histidine residue undergoes autophosphorylation. There are two response regulators called CheB and CheY. There is a transfer of a phosphoryl group to their conserved aspartate residue from CheA. CheY subsequently interacts with the flagellar switch protein called FliM. This induces the switching in flagellar direction from counter-clockwise to clockwise.
This clockwise rotation upsets the entire flagella bundle and causes it to break up. The result is that the bacterium “tumbles.” This means that bacteria are able to re-direct their course and repeatedly re-evaluate and adjust their bearings in response to environmental stimuli such as food or poisons.
As for the other response regulator I mentioned, CheB, what does it do? This is where it gets really good. When CheB is activated by the histidine kinase CheA, it operates as a methylesterase. This means that it actively removes methyl groups from glutamate residues on the receptor’s cytoplasmic surface. Meanwhile, another protein (called CheR) actively adds methyl residues to these same glutamate residues: that is to say, it works as a methyltransferase.
At this point the engineering shows a stroke of genius. If the stimulus is at a high level, there will be a corresponding decline in the level of phosphorylation of the CheA protein: and, as a consequence, of the response regulators CheY and CheB as well. Remember that the role of CheB is to remove methyl groups from glutamate residues on the receptor’s cytoplasmic surface. But now, phosphorylated CheB is not available and so this task is not performed. The degree of methylation of the MCPs will thus be raised. When the MCPs are fully methylated, the cell will swim continuously because the MCPs are no longer responsive to the stimuli.
This entails that the level of phosphorylated CheA and CheB will increase even when the level of attractant remains high, and the cell will commence the process of tumbling. But now, the phosphorylated CheB is able to demethylate the MCPs, and the receptors are again able to respond to the attracting chemical signals. In the case of repellents, the situation is similar — except that it is the least methylated MCPs which respond least while the fully methylated ones respond most. This kind of regulation also means that the bacterium has a memory system for chemical concentrations from the recent past and compares them to its currently receiving signals. It can thus detect whether it is moving towards or away from a chemical stimulus.
A diagrammatic illustration of this system is to be found in the figure below:
Proton Motive Force
A proton motive force drives the rotation of the bacterial flagella. Electrochemical energy is converted into torque via an interaction between the stator and the rotor. Torque is then transmitted from the C-ring by the MS ring to the rod and then to the hook region. From there it is transferred to the propeller (filament). When the filament rotates, the torque is converted into thrust, allowing the bacterial cell to move.
The rotary engine is located at the flagellum’s anchor point on the inner membrane. A flow of protons across the membrane, resulting from a concentration gradient, drives the motor. Protons are transferred across the membrane by the rotor, and the rotor is turned in the process. The speed at which the motor turns can be increased or decreased as a direct result of changes in the strength of the proton motive force.
But it only gets better. An interesting 2008 study by Blair et al. demonstrated that an operon required for the biosynthesis of the biofilm matrix in Bacillus subtilis also encodes a molecular clutch, called EpsE. When associated with a flagellar basal body, EpsE disables the biological motor by disengaging the drive train from the power source! Those hungry for further reading are invited to read Howard Berg’s 1995 paper on Torque generation in the bacterial flagellar rotory motor.
Flagellar Assembly
The processes of flagellar assembly, and its regulation, is my personal unparalleled favourite area of study in the discipline of microbiology. For an informative documentary on the assembly of this nanomachine, I highly recommend viewing this video.
The self-assembly of the bacterial flagellum requires the co-ordinated expression of more than 60 gene products (which encode for both structural and regulatory proteins). Bacteria are intriguingly able to detect the stage of progress with respect to flagellar assembly, and subsequently use this information in the co-ordination of gene expression.
The mechanisms of flagellar assembly are so complex and so sophisticated that justice can hardly be done to them here. Interested readers who desire a fuller discussion of these mechanisms are advised to read a 2002 paper, authored by Phillip Aldrige and Kelly T Hughes, appearing in Current Opinion in Microbiology. In addition, a technical book, Pili and Flagella: Current Research and Future Trends features a detailed discussion of this topic. The book is typically priced at £150/$310, but a significant portion of the book may be accessed via Google Books. The book discusses studies of flagellar mutants that fail to complete their flagella. In so doing, they discern which proteins are absolutely indispensible to flagellar assembly, and which are not — something any ID proponent should be interested in.
Flagellar assembly begins in the cytoplasmic membrane, progresses through the periplasmic space, and finally extends outside the cell. As previously alluded to, the flagellar apparatus consists of two main parts: the secretion system and the axial structure. The main components of the axial structure are FlgG for the rod, FlgE for the hook, and FliC for the filament. All of these assemble with the help of a cap protein (FlgJ, FlgD and FliD respectively). Of those, only FliD remains at the filament’s tip — the other two are not present in the finished product. Other components of the axial structure (FlgB, FlgC and FlgF) connect the rod and MS ring complex. The hook and the filament are connected by FlgK and FlgL
The MS ring complex is the structural foundation of the apparatus. When the C ring and C rod attach to the M ring (at its cytoplasmic surface), the complex begins to secrete flagellar proteins.
With the assistance of a cap protein (FlgJ), the rod structure is constructed through the peptidoglycan layer. But its growth is terminated when it reaches the outer membrane (which represents a physical barrier such that it cannot pass through unaided). But not to worry! The outer ring complex cuts a hole in the membrane. The hook then begins to grow beneath the FlgD scaffold until it reaches approximately 55nm in length. When the hook reaches this critical length, the substrates which are being secreted switches from the rod-hook mode to flagellin mode. FlgD is then replaced by HAPs (hook associated proteins) and the filament continues to grow — note that this can only take place in the presence of FliD (the cap protein), otherwise the flagellin monomers are lost.
The Regulation of Flagellar Assembly
The energy costs of assembling flagella make these nanomachines expensive systems to run. This entails the necessitude for systems and mechanisms to regulate the co-ordination of assembly. The flagellar system of Salmonella (the paradigm organism for this system) possesses three classes of flagellar promoters which are organised into a transcriptional hierarchy. The Class I promoter drives the expression of the enteric master regulator, FlhD4C2. In association with the sigma factor, σ70, this master regulator turns on the Class II promoters which are responsible for the gene expression of the hook-basal-body subunits and its regulators, including σ28, and its anti-sigma factor, FlgM. σ28 is required for the activation of the Class III promoters. This class of promoters is responsible for the expression of flagellin monomers, the chemotaxis system and the motorforce generators. Interaction with its anti-sigma factor (FlgM), thus keeping it away from the RNA polymerase holoenzyme complex, inhibits the activation of σ28 prior to the completion of the Hook Basal Body. When the Hook Basal Body is finished, the FlgM is secreted through the flagellar structure via the Type III Secretion Apparatus substrate specificity switch. This means that the Class III promoters can now be activated by their sigma factor (σ28) because its anti-sigma factor (which represses its action) has been removed. This allows the flagellum to be completed.
This brief overview of flagellar assembly has barely scratched the surface of this intricate process. Nothing will please me more than if, after having read this description, at least a few readers reach for the relevant literature to find out more.
The Bacterial Flagellum — A Paradigm for Design?
In writing the above descriptions, I hoped to treat the educated layperson to a flavour of the magnificence of this rotary motor. As previously stated, nothing will bring me more satisfaction than having provided someone with the inspiration to pursue further information. The question does arise, however, as to whether this system bears the hallmarks of design — or is it, as so many biologists maintain, merely the product of blind, purposeless and impersonal natural processes of chance and necessity? To me, the answer is very clear. This system certainly appears to be designed technology. It certainly appears to bear the hallmarks of design logic. And it does seem that the burden of proof must, at this present time, lie with he who rejects this proposition — that is, he who ascribes this system to blind natural processes. The handwaving gestures of Kenneth Miller, who asserts that the Darwinian explanation for this system is more appropriate in light of the presence of the Type III Secretion System, can no longer be considered a satisfactory rebuttal to the design postulate. Such gestures may, at first glance, appear convincing to the uninitiated, unfamiliar with the complexities of these systems. But when one examines the molecular structure and orchestration of these systems, one loses satisfaction with the vague discussions of protein sequence homologies. Kathryn Applegate and Ard Loius seem to be under the impression that this assembly process parallels the processes of Darwinian evolution — I will allow readers to judge for themselves on that one.
As has been pointed out by Stephen Meyer and Scott Minnich, there is an even more fundamental challenge to the standard Darwinian line with regards this system. Firstly, the phylogenetic evidence is strongly suggestive that the T3SS is an evolutionary biproduct of the flagellar system, rather than the other way round (see, for instance, here). Secondly, if flagellum biosynthesis were to be expressed simultaneously with the Yop T3SS, flagellin monomers would likely be exported out the needle-like structure as well as the flagellar basal body, potentially limiting the efficiency of both systems. Meyer and Minnich argue in their paper that the potential for cross-recognition between Type III exported proteins in the same cell explains why the segregation of these systems by specific environmental cues is necessary. Expression of a flagellum under host conditions would result in a loss of polarised secretion of Yop proteins into the cells of the host. Flagellin is also a potent cytokine inducer – display of flagellin to the macrophages by direction injection via the Ysc secretin would strongly countermand the Yersinia’s anti-inflammatory strategy.
Again, interested readers who are hungry for additional material are directed to this video, which features microbial geneticist, Scott Minnich, presenting an insightful lecture on this remarkable motor.