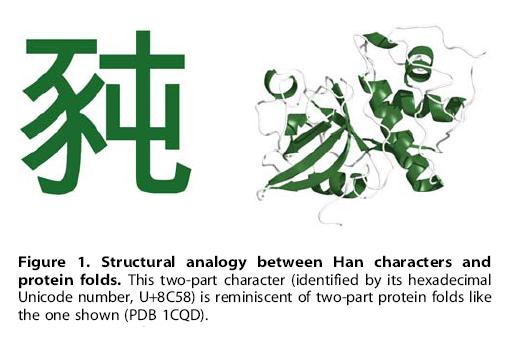
Uncommon Descent holds that ...
Materialistic ideology has subverted the study of biological and cosmological origins so that the actual content of these sciences has become corrupted. The problem, therefore, is not merely that science is being used illegitimately to promote a materialistic worldview, but that this worldview is actively undermining scientific inquiry, leading to incorrect and unsupported conclusions about biological and cosmological origins. At the same time, intelligent design (ID) offers a promising scientific alternative to materialistic theories of biological and cosmological evolution — an alternative that is finding increasing theoretical and empirical support. Hence, ID needs to be vigorously developed as a scientific, intellectual, and cultural project.