 |
Dr. Ann Gauger argues that the hypothesis of common descent fails to account for the origin of the mammalian placenta, in an ENV article titled, The Placenta Problem (June 17, 2016). As we’ll see, the evidence she puts forward proves precisely the opposite: common descent is the only hypothesis which explains the facts, without resorting to ad hoc suppositions.
I’d like to begin with a confession. When I read Dr. Gauger’s article on the origin of the placenta, my first reaction was: “Whoa.” It appeared that Dr. Gauger had made a very strong case against the hypothesis of common descent. But then I did some more reading, and after looking at the evidence which Professor Joshua Swamidass kindly forwarded to me, I came to realize that my initial impression was mistaken. For laypeople like myself, Carl Zimmer’s highly readable and refreshingly jargon-free article, Mammals made by viruses (Discover magazine, February 14, 2012) provides an excellent overview of the evidence, from an evolutionary standpoint. Readers with a background in biology may find this 2015 article by Imakawa et al. more interesting.
But first of all, let’s examine what Dr. Gauger has to say:
According to the theory of common descent, all true mammals are supposed to have descended from a common ancestor with a placenta. This is a trait common to all mammals. However, it has been a puzzle for some time that placentas differ in the form they take among different mammalian clades.
As an Australian, I feel bound to point out that Dr. Gauger’s description of placental mammals as “true mammals” is biologically incorrect. For example, monotremes (such as the Australian platypus) are mammals that lay eggs, and they don’t have a placenta. Marsupials (such as the kangaroo, the koala and the North American opossum) are mammals that carry their young in a pouch. They also do not have a placenta. Nevertheless, they are true mammals.
I’m also mystified by Dr. Gauger’s assertion, later on in her article, that “in 2015 a functional syncytin was found in several marsupials, extending the presence and essential function of the protein to all placental mammals examined.” I’m quite sure Dr. Gauger is perfectly well aware that marsupials are not placental mammals.
But let us continue with Dr. Gauger’s case against common descent (bolding is mine – VJT):
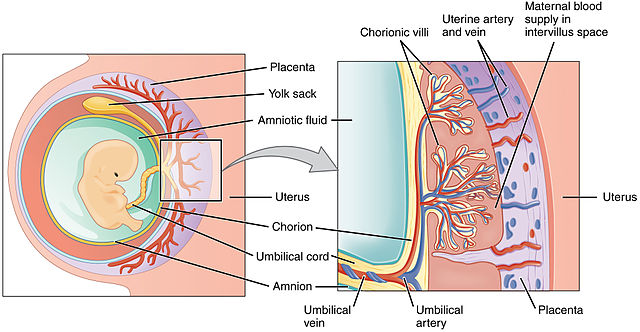
In the year 2000, French researcher Thierry Heidmann and coworkers found that genes derived from endogenous retroviruses (ERVs) appear to have been coopted to perform an essential role in placental formation. These genes, which resemble the ERV envelope gene env, make a protein that originally promoted fusion of the virus with its host cell’s membrane, but now acts to promote fusion of membranes between the embryo and the lining of the uterus. These “repurposed” proteins are called syncytins. They are essential for placental formation, yet are of independent origin in different kinds of mammals — primates have one kind, mice another, rabbits, cows, and carnivores yet others. They are clade-specific. In fact, in 2015 a functional syncytin was found in several marsupials, extending the presence and essential function of the protein to all placental mammals examined. All syncytins are lineage-specific, meaning that each mammalian clade has its own syncytin, with a unique sequence and location in the genome. They must have inserted themselves (or been placed there) after the separation of the mammals into different clades! This means there must have been multiple independent acquisitions of these syncytins to participate in an essential process that is common to all mammals. Why should there be unique syncytins in each clade?
What we have to explain is the unique and independent group-specific cooption of syncytins for a function that is essential for placental development, a feature common to all mammalian groups. Six independent origins for the placenta! There is no evidence of a grand ancestral syncytin shared by all groups that was later replaced by other syncytins, so the common descent explanation of the placenta in mammals fails…
Rather than postulating six independent, random capture events in placental development, they [Lavialle et al., the authors of a review paper on syncytins] are now postulating at least one more, a founding syncytin leading to a primitive placenta, then the other syncytins to replace that one in each lineage. Each replacement must have had a clear selective advantage as time went on to make the replacement possible, and each must be the outcome of a random series of events. To say it again, the common descent prediction is that there must have been a founding syncytin in the first mammal with a placenta, or something else that functioned in syncytin’s place, in order for the primitive placenta to arise and subsequently be passed to all mammalian clades. For which there is no evidence, and may never be.
Can common descent explain the unexpected observation of six independent origins for the placenta? No. Could it predict it? No….
In considering these alternative explanations, ask yourself, how likely is it that a retrovirus would infect, invade the germ line (the cells that make eggs and sperm), then insert itself at random in locations in the genome that are expressed in the developing embryo or primitive uterus at the proper time, then promote fusion of membranes to permit the formation of a placenta, with all this happening at least six separate times in the six lineages tested so far? We should also make clear, expressing a syncytin by itself is unlikely to be enough to make a placenta, which is a complex organ requiring interactions between mother and embryo, and the ability to exchange nutrients and oxygen.
Dr. Gauger has put together what appears to be a very powerful case. So how would a proponent of common descent answer it?
Stages in the evolution of the mammalian placenta
What we need to keep in mind is that the evolution of the mammalian placenta would have required a large number of steps. At the present time, we do not know precisely how these steps would have been implemented, on a genetic or anatomical level. Nor do we know exactly how many steps would have been required. However, Professor Swamidass has outlined the major changes that would have taken place.
In the ancestors of today’s mammals, egg-laying in a wet environment would have been replaced by the appearance of parchment-shelled eggs. Next, egg wetting (or the supply of water to parchment-shelled eggs, which can rapidly lose moisture in a hot, dry environment) would have evolved, followed by the appearance of an early placenta. After that, the placenta would have undergone further refinement, with better separation between fetal/maternal cells so as to enable longer gestation, leading finally to a modern placenta.
Of the changes listed above, the first three are the most critical. Professor Swamidass suggests that the early steps in mammalian evolution, shared by all mammals, were either low probability or highly contingent on the exact genetic, functional, and environment of earliest mammals. For example, parchment eggs that required egg wetting might have been the low probability event that enabled the whole sequence.
Syncytins, on the other hand, relate to the penultimate phase (better separation between fetal/maternal cells), which is one of the easiest steps in the evolution of the placenta. Syncytins are just one of many ways of accomplishing better separation between fetal and maternal cells, so as to enable longer gestation. There are many ways of doing this, but acquiring a gene from a syncytal virus is a particularly easy way. It is not at all surprising that this happened several times, in different lineages of mammals.
Would the insertion of a retrovirus in a mammal’s genome have been an extremely unlikely event?
 |
Dr. Ann Gauger contends that the capture of a retrovirus containing a gene capable of encoding a protein required for the formation of a placenta, coupled with the insertion of this retrovirus into an appropriate location in a mammal’s genome, where it can be expressed at the right time, would have been a staggeringly unlikely event:
In considering these alternative explanations, ask yourself, how likely is it that a retrovirus would infect, invade the germ line (the cells that make eggs and sperm), then insert itself at random in locations in the genome that are expressed in the developing embryo or primitive uterus at the proper time, then promote fusion of membranes to permit the formation of a placenta, with all this happening at least six separate times in the six lineages tested so far?
However, Professor Swamidass has informed me that it is actually very common for mammals to be infected by viruses with syncytins, and that it is also very common for them to insert themselves into the genome. The only rare part would inserting into the right place in the genome, so that the genetic expression is right. Given that we only need to have this happen once every several million years, Swamidass sees no difficulty here.
Now, perhaps some readers may disagree with Professor Swamidass’s reasoning here. Fine. But if you disagree, then please, let us see your calculations – even if they are merely calculations of the “back-of-the-envelope” variety.
While we’re on the topic of syncytin genes, allow me to quote from a perspicacious remark by a commenter named Evolve, in a recent thread on Uncommon Descent (bolding is mine – VJT):
Ann Gauger thinks she has nailed the case against common descent through the example of syncytins because they were not inherited by all placental mammals from their supposed common ancestor. She says that different syncytins, with no common origin, were inserted by a designer into specific mammalian lineages at different time points. But her logic is utterly flawed because syncytins did not pop into existence from nowhere one fine morning. They are viral proteins and viral infections are very common in all vertebrates including mammals. It is also common for viral genes to integrate into the host organism’s genome (all mammalian genomes sequenced thus far are littered with tons of viral gene remnants, which emphatically prove this point). Occasionally, some viruses infect the germline too and, when this happens, viral genes can get passed down to the next generation. Different syncytins were inherited by different mammalian lineages from such infections.
Syncytins perform two main functions for the virus – they suppress the host’s immune response and they facilitate fusion of the viral envelope with the host cell. When syncytins were acquired by the host organism, both these functions were co-opted for different purposes. The immune suppressing role was used to prevent rejection of the foetus by the mother, while the fusogenic (fusion-promoting) role was used to form the placenta – a tissue that results from the fusion of maternal and fetal cells.
Syncytins are not mysterious, magic genes Ann Gauger’s designer pulled out of thin air. Their origin and co-option for different purposes by mammals occurred through well-understood processes. As such, a designer is not the most parsimonious explanation, and it is unnecessary and redundant.
Is the placenta likely to have been designed?
So, was the mammalian placenta designed? My own answer to this question would be: “Yes, but design is not an all-or-nothing affair.” I would guess that Intelligent Design was required during the first three phases in the evolution of the placenta (discussed above), but not during subsequent phases. I may of course be wrong here; but if you think I am wrong, please provide me with some solid reasons. The capture of a founding retroviral env gene, and its subsequent replacement by new env-derived syncytin genes, doesn’t sound like an intelligently designed process to me. Still, I might be mistaken.
Do advocates of common descent postulate six different origins of the placenta?
No. Advocates of common descent do NOT postulate “six independent origins for the placenta,” as Dr. Gauger claims in her article. That is a misunderstanding. Taken by itself, the hypothesis of common descent is completely agnostic as to whether the different placentas we find in mammals have a single, unified origin in the ancestor of modern-day placentals, or multiple, independent origins in different lineages of placental mammals.
Professor Swamidass points out that there are two hypotheses that could explain the data. On the one hand, it could be a case of convergent evolution, where syncytin is acquired independently by different lineages of placental mammals. Alternatively, there may have been only one origin of the placenta, followed by six independent capture events. In order for this to happen, there would have to be a “baton pass” mechanism, enabling each lineage to replace the original syncytin that would have been present in the ancestor of modern placentals. However, the key point, as we’ll see below, is that while the genes required for the formation of a placenta differ across various lineages of mammals, they follow a lineage-specific pattern, which is consistent with common descent. It is very easy to imagine patterns in the data that would contradict common descent, but we do not see these patterns.
Readers may be wondering how a “baton pass” mechanism would work. That’s a reasonable question. Professor Swamidass has kindly forwarded me a copy of a 2015 paper titled, Baton pass hypothesis: successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution, by Kazuhiko Imakawa, So Nakagawa, and Takayuki Miyazawa (Genes to Cells, Volume 20, Issue 10, October 2015, pages 771–788). The paper’s abstract puts forward the authors’ hypothesis:
The syncytin genes so far characterized are known to be endogenized to the host genome only within the past 12–80 million years, more recently than the appearance of mammalian placentas, estimated to be 160–180 million years ago. We speculate that ERVs [endogenous retroviruses – VJT] including syncytin-like gene variants integrated into mammalian genomes in a locus-specific manner have replaced the genes previously responsible for cell fusion. We therefore propose the ‘baton pass’ hypothesis, in which multiple successive ERV variants ‘take over’ cell-fusion roles, resulting in increased trophoblast cell fusion, morphological variations in placental structures, and enhanced reproductive success in placental mammals.
 |
The authors also propose a highly specific mechanism for how this baton pass might work:
As in an actual baton pass, the two (or more) genes briefly share the same function in the genome of a given host species. Eventually however, the newly acquired gene ‘takes over’, becoming more prominent as the previous gene’s relative importance subsides. The previous gene may be lost or co-opted for another function such as an immunosuppressive property, also essential for mammalian pregnancy (Esnault et al. 2013; Lavialle et al. 2013). In this hypothesis, gene evolution through ERV integration could proceed more quickly than in more conventional models of gene evolution, because the transcription of tissue-specific and/or neighboring genes may regulate ERV gene expression (Yu et al. 2002; Muroi et al. 2009; Nakaya et al. 2013). For the ERVs that function in reproductive processes, the integration of ERVs must be locus-specific because they could be transcribed with their own LTRs or along with placenta-specific genes (Dewannieux & Heidmann 2013; Nakaya et al. 2013). In the baton pass hypothesis, acquisition of ERVs still requires changes in chromosome/genome structures, as it takes over and modifies the preexisting function such as trophoblast cell fusion among mammalian species, enabling novel evolutionary changes.
Under this model, the original placental would have had a syncytin, but the “baton pass” mechanism would have subsequently enabled different lineages of placental mammals to genetically drift to using new and different syncytins.
Now, Dr. Gauger is perfectly entitled to point out that there is, at the present time, no evidence for a founding syncytin in the first mammal with a placenta. Fair enough. But as we’ll see below, the hypothesis of common descent makes a singular prediction about the genes that play a key role in the formation of placentas, which no other hypothesis makes. This bold prediction makes the hypothesis of a founding syncytin a reasonable one. Whether it should prove to be true or false, the “baton pass” theory is more than mere speculation: it is a plausible hypothesis.
What singular predictions does the hypothesis of common descent make?
As Professor Swamidass has pointed out above, the hypothesis of common descent does not predict a single, common origin for placentas, or for the genes involved in their development. What it does predict is that the genes that play a key role in the formation of placentas should always be in the same location, in the genomes of genetically similar mammals (e.g. primates). Of course, the genes that play a key role in the formation of the placenta may be located in totally different positions in the genomes of different lines of mammals (e.g. Old World primates vs. rats and mice). And this is precisely what we find. Even though they all originated from a similar biochemical mechanism, being derived from endogenous retroviruses that became embedded in mammals’ genomes, the specific DNA insertion sites for these placental genes are different, in different lines of mammals, and they line up by clade. Dr. Gauger herself acknowledges as much:
In the year 2000, French researcher Thierry Heidmann and coworkers found that genes derived from endogenous retroviruses (ERVs) appear to have been coopted to perform an essential role in placental formation. These genes, which resemble the ERV envelope gene env, make a protein that originally promoted fusion of the virus with its host cell’s membrane, but now acts to promote fusion of membranes between the embryo and the lining of the uterus. These “repurposed” proteins are called syncytins. They are essential for placental formation, yet are of independent origin in different kinds of mammals — primates have one kind, mice another, rabbits, cows, and carnivores yet others. They are clade-specific. (Bolding mine – VJT.)
Precisely. This is just what the hypothesis of common descent would predict.
Professor Swamidass expresses the point very succinctly:
CD [Common descent] does not predict that syncytin arises only once. CD itself just predicts that each specific event will fit into the tree.
Likewise, CD does not predict one common origin for placentas. It predicts that the genetics will be well nested in clades, which it is.… (Bolding mine – VJT.)
What would really upset proponents of common descent is if we were to find that a few species of primates (e.g. human beings) possessed the version of syncytin found in rats and mice, instead of the version found in most Old World primates. Alternatively, if the genes involved in the formation of the placenta in human beings were in a different location from the corresponding genes in other Old World primates, that would also tell heavily against the theory of common descent.
As Professor Swamidass has pointed out, the reason why these results lend such solid support to the theory of common descent is that they demonstrate that on a genetic level, there are even more types of placentals than scientists had previously believed, on the basis of purely anatomical comparisons. What’s more, these genetic types cluster perfectly, just as the hypothesis of common descent would predict.
I should add that from a common design (as opposed to common descent) perspective, it would have been very easy for the Designer to have given evidence against common descent in our genomes. For instance, the Designer could have given human beings placentas a genetically different placenta. He could have inserted our syncytins in a location in our genome which is inconsistent with the hypothesis of common descent. Alternatively, He could have given us entirely different syncytins. There’s no biological constraint which prevents the Designer from doing either of these things. But He didn’t.
Carl Zimmer summarizes the evidence that humans and other primates share a common ancestry in his article, Mammals made by viruses (Discover magazine, Februaey 14, 2012):
It turned out that syncytin was not unique to humans. Chimpanzees had the same virus gene at the same spot in their genome. So did gorillas. So did monkeys. What’s more, the gene was strikingly similar from one species to the next. The best way to explain this pattern was that the virus that gave us syncytin infected a common ancestor of primates, and it carried out an important function that has been favored ever since by natural selection. Later, the French virologist Thierry Heidmann and his colleagues discovered a second version of syncytin in humans and other primates, and dubbed them syncytin 1 and syncytin 2.
There is no known principle that explains all of the anatomical and genetic features of human placentas, except common ancestry with apes. That’s why the strong genetic similarity between chimps and humans is evidence for common descent.
Finally, Professor Swamidass explains why the genetic evidence for common descent from the study of placentas is much stronger than the anatomical evidence. When you look at anatomy, he says, you can see similar patterns in animals belonging to the same group, but this often requires very careful study, and anatomical comparisons between animals invariably involve an element of subjectivity. Genetic comparisons, however, are not subjective. They are very clear. The genetic data is 100% consistent with common descent. It could have contradicted it, but it didn’t. Only common descent explains why the genetic data has to cluster in clades. On a common design hypothesis, it might, but it doesn’t have to.
At this point, I’d like to invoke an explanatory principle commonly used by scientists:
If hypothesis A predicts that X must occur, and no other known hypothesis makes that prediction, and X occurs, then hypothesis A is a better explanation of X than other hypotheses, and (ceteris paribus) more likely to be true.
Common design doesn’t explain why the genetic data has to cluster in clades. It only says that it might – but then again, it might not. That’s not a prediction. Only common descent unambiguously predicts that it will cluster in clades. That’s what makes it a better hypothesis.
It may also be objected that a theory of common descent, taken by itself, supplies no mechanism for evolutionary change. However, common descent is best regarded as an explanatory framework. It is a foundation, to which we can append more precise claims about mechanisms, in order to form a testable hypothesis. Adding the neutral theory of evolution generates a hypothesis with a good track record of correctly predicting/modeling the data.
Finally, I should add that in very rare cases, the theory of common descent may need to be supplemented by a theory of Intelligent Design, to help it surmount any probabilistic hurdles involved in the evolution of the placenta that would have been insuperable via unguided natural processes. However, the existence of these hurdles has yet to be demonstrated, so we should all keep an open mind.
Does Ann Gauger’s hypothesis of convergent design account for the origin of the placenta?
At this point, critics of Dr. Gauger may be wondering what her alternative to common descent is. Dr. Gauger has a ready response to this question:
Common design has an explanation, but not one that will be palatable to my interlocutors. The designer used the same idea six different times to produce the same outcome in six different “designs” (clades). That’s another way of saying all these clades have the same outcome, the placenta, but achieved by independent uses of the same idea…
Convergent design is to be expected under the design hypothesis because the designer is not constrained by an evolutionary tree. He can reuse ideas that work in one setting in a different place. In fact, he can mix and match his methods to get to any outcome he wants. I am thinking of echinoderms (sea stars and sea urchins) that look alike as adults but get there by very different developmental paths, or two very different animal groups that come up with similar molecular solutions to create a new function, echolocation…
However, Professor Swamidass finds this line of reasoning inconsistent. On the one hand, Dr. Gauger appears to be arguing that homology and synteny constitutes evidence that the Designer is copying from a common design to make new organisms. In other words, when similar DNA does very different things, this counts as evidence of design. But on the other hand, Dr. Gauger also argues that multiple DNA coding solutions that do similar things are designed, too. In other words, when totally different DNA blueprints are used to generate very similar solutions, somehow this also counts as evidence of design. There’s a name for this kind of logic: heads, I win; tails, you lose.
Unfortunately, Dr. Gauger makes no attempt to explain why the Designer sometimes uses repeated DNA blueprints for different purposes, and why He sometimes uses different DNA blueprints for the same purpose. What design principle is at work here? And why don’t the platypus and the opossum have a placenta? No answer is provided.
By contrast, the hypothesis of common descent explains:
(i) the full range of genetic and anatomical differences and similarities we see in placental mammals;
(ii) why syncytins could have arisen multiple times (it’s a high-probability way to solve one step in the path to a modern mammalian placenta); and
(iii) why the genetically related placentals group into nested clades (this is an automatic prediction of common descent, if we assume that the process generating nested hierarchies of classification in placental mammals is a memoryless Markov chain).
Taken together, all of this data constitutes very powerful evidence for common descent.
To sum up: we see several patterns that are easily explained by the hypothesis of common descent. None of them are explained by a consistent design principle.